“Especially in the context of NALFIAs, the PCR is may be the most underrated
amplification technique with the biggest untapped potential.“
Polymerase Chain Reaction & Lateral Flow
Polymerase Chain Reaction – A Short Summary
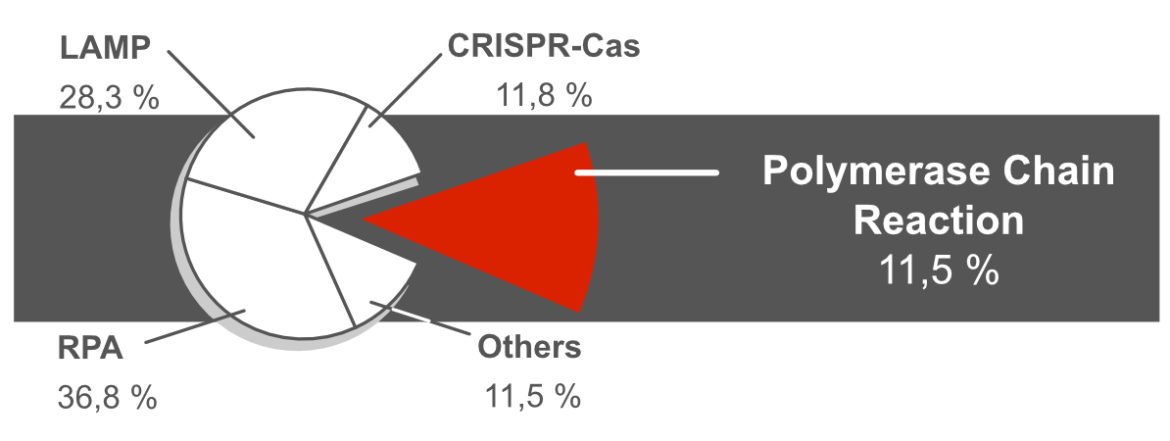
Polymerase Chain Reaction (PCR) is the oldest, best understood and most diverse DNA amplification technique. However, it plays a rather subordinate role in combination with lateral flow based readout formats. Only about 11% of the NALFIA related publications refer to the use of PCR. The reasons for this are frequently mentioned in scientific publications. Polymerase Chain Reaction is often characterized as: laborious, time-and cost-intensive, needs special equipment and highly trained personnel. Here, various points are explained and discussed in order to critically question these arguments. This is a little teaser:
History of the Polymerase Chain Reaction
The concept of the polymerase chain reaction was initially described by Kary Mullis in the early 1980s. About ten years later, PCR has become one of the most important tools in molecular biology. In 1993, Kary Mullis received the well-deserved nobel prize in chemistry for inventing PCR. Today, more than 40 years after the first publications of this DNA amplification technique, PCR forms the basis for countless molecular biological methods and is used in manyfold in everyday life. A very concise example is the field of molecular diagnostics, which go far beyond human medical issues.
Invention
PCR method was invented by Kary Mullis
Landmark Paper
First description of the PCR-technique by Randall Saiki et al. (Science)
Themal Cycler
First commercially available PCR machine introduced
Taq Polymerase
Heatstable DNA-polymerase from Thermus aquaticus patented by Mullis
RNA-Detection
Combination of reverse transcription and PCR for the detection or RNA (RT-PCR)
Nobel Prize
Micheael Smith & Kary Mullis received nobel prize in chemistry
quantitative PCR
Quantitative Analysis with Real-Time PCR
PCR & Lateral Flow
One of the first descriptions for the combination of PCR & Lateral Flow Analysis by Kozwich et al. (Appl. Environ. Microbiol.)
Invention
PCR method was invented by Kary Mullis
Themal Cycler
First commercially available PCR machine introduced
RNA-Detection
Combination of reverse transcription and PCR for the detection or RNA (RT-PCR)
quantitative PCR
Quantitative Analysis with Real-Time PCR
Landmark Paper
First description of the PCR-technique by Randall Saiki et al. (Science)
Taq Polymerase
Heatstable DNA-polymerase from Thermus aquaticus patented by Mullis
Nobel Prize
Micheael Smith & Kary Mullis received nobel prize in chemistry
PCR & Lateral Flow
One of the first descriptions for the combination of PCR & Lateral Flow Analysis by Kozwich et al. (Appl. Environ. Microbiol.)
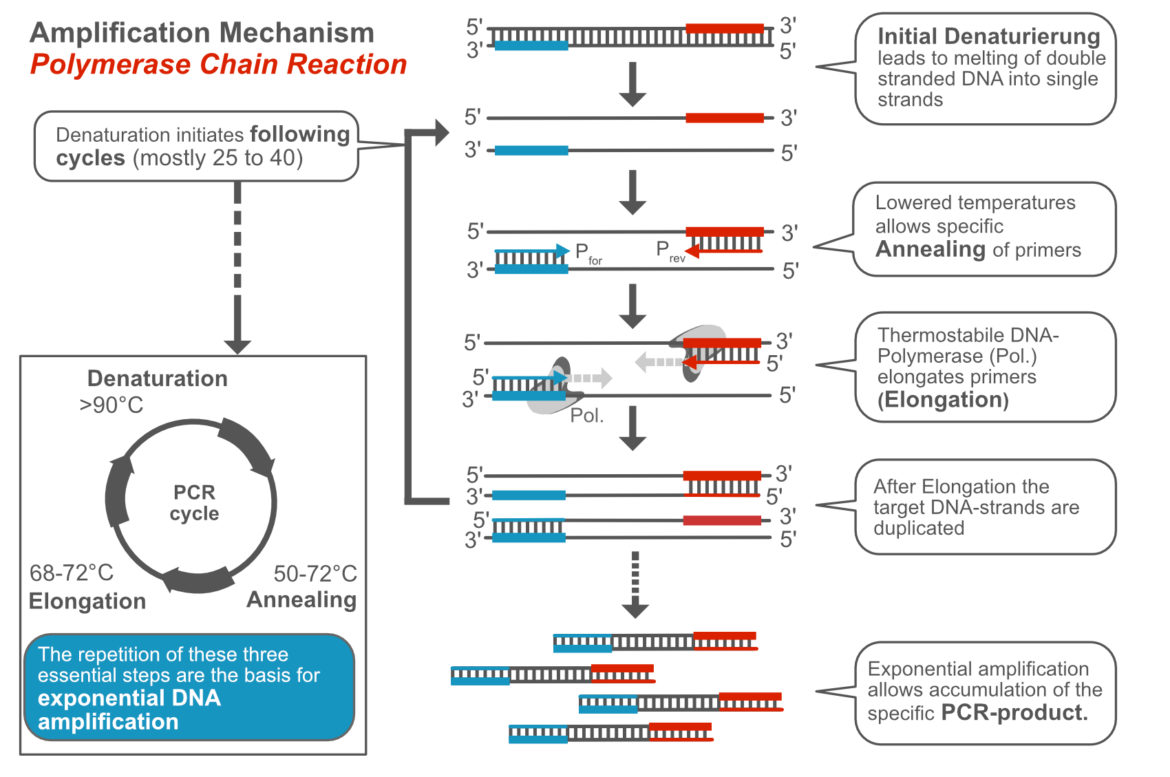
General Mechanism of the Polymerase Chain Reaction
The mechanism of the Polymerase Chain Reaction has been described over and over again. We try to keep it as simple as possible. First of all, you have to know what the PCR is. The Polymerase Chain Reaction is an enzyme-driven in vitro DNA-amplification technique, which allows the specific and sensitive detection of low abundant template DNA. The Polymerase Chain Reaction is subdivided into three characteristic, temperature-dependent steps:
- Denaturation – melting double stranded template DNA into single strands (>90°C)
- Annealing – specific binding of primers to ss-template-DNA (50-70°C)
- Elongation – DNA-synthesis is catalized by heat stable DNA-polymerase, which elongates primers (68-72°C)
The cyclical repetition of these three essential steps allows exponential amplification a defined DNA fragment. Within 30 cycles, more than 1 billion DNA copies can be generated from a single template molecule. The following video „from 1 to 1 billion DNA copies – PCR explained in one minute“ shows the principle of PCR.
Modification of a PCR for Lateral Flow based Readout
In order to visualize a PCR product on a universal Lateral Flow Device (LFD), labels must be incorpotated somehow into the double strand. Various labeling strategies are applicable, which should be carefully considered before starting assay development. The labeling strategy is directly linked to the selected PCR methodology. In addition to the incorporation of the labels, other methodological modifications can be used, which can be particularly important for PCR-LFA. An example for this is the PCR temperature protocol. It is an important tool within a PCR development project and can therefore make a significant contribution to specificity and sensitivity of the assay.
Terminal labeling of PCR primers may be the easiest and most intiutive way to detect a PCR product on a universal Lateral Flow Device. Labeled primers are efficiently incorporated into amplicons by most DNA-polymerases during the amplification reaction. These short, dual labeled PCR products can be detected 100 to 1000 times more sensitive with Lateral Flow Analysis compared to standard agarose gelelectrophoresis.
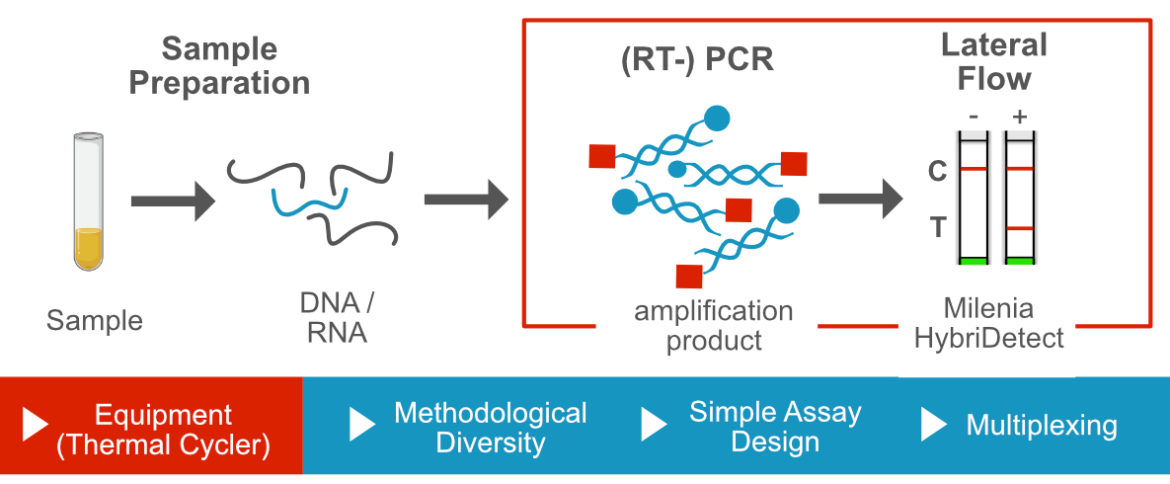
The temperature protocol of a PCR should be understood as a characteristic and important tool. Compared to isothermal amplification methods, the interaction of the PCR composition and the temperature profile can be used in advance for the development of an excellent detection performance.
Methodological Variations of Polymerase Chain Reaction
PCR is probably the most multifaceted and flexible DNA amplification technique. PCR-LFA can particularly benefit from this. The choice of the right PCR method for the application should be considered carefully. The conditions under which the method should finally be used are absolutely decisive for the choice of the appropriate detection technique.
Advantage: Multiplexing
Multiplexing is the ability to amplify multiple, defined products in one reaction. PCR in particular is characterized by the fact that it allows a strong multiplexing. The simple assay design in combination with the multiplex compatibility underscores the value of the Polymerase Chain Reaction in general and especially in the field of Point-of-Care diagnostics.
Learn more about (PCR) Multiplexing & Lateral Flow
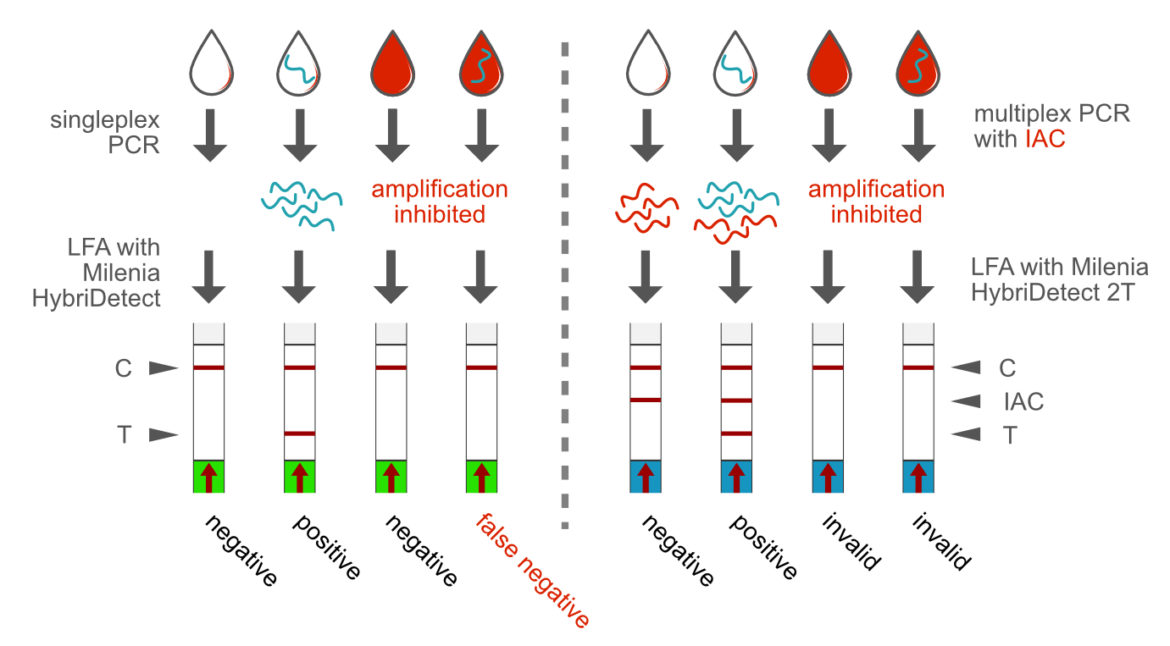
Direct PCR and Internal Amplification control (IAC)
A wide variety of sample matrices can be used in PCR, if a suitable and robust enzyme system has been selected. This is the prerequisite for the direct application of a biological material in a PCR, saving the demand for a DNA or RNA extraction prior to do the PCR reaction. This is an important feature for the development of point-of-care compatible rapid tests. Direct analysis from blood is just one example of practical relevance. The direct PCR format can benefit from the multiplex ability by implementing an internal amplification control (IAC). This methodological approach allows a clear differentiation between negative results and matrix-dependent PCR inhibition. The Milenia HybriDetect 2T allows rapid evaluation of a duplex PCR.
Advantage: Specificity – Detection of SNPs
 PCR is often used in molecular diagnostics when extremely specific detection is required. An example for this is the identification of single nucleotide polymorphisms (SNPs). A SNP is a substitution of a single nucleotide at a certain position in a genome. Mostly SNPs are detected by special sequencing-, array- or PCR techniques. But the combination of PCR and lateral flow analysis is particularly suitable for a rapid and Point-of-Care compatible SNP analysis. For example, the amplification-refractory mutation system (ARMS) – PCR has proven to be a useful tool for detection of SNPs over decades. We could also demonstrate, that simple hybridization can be very useful for genotyping or multiplex SNP-detection.
PCR is often used in molecular diagnostics when extremely specific detection is required. An example for this is the identification of single nucleotide polymorphisms (SNPs). A SNP is a substitution of a single nucleotide at a certain position in a genome. Mostly SNPs are detected by special sequencing-, array- or PCR techniques. But the combination of PCR and lateral flow analysis is particularly suitable for a rapid and Point-of-Care compatible SNP analysis. For example, the amplification-refractory mutation system (ARMS) – PCR has proven to be a useful tool for detection of SNPs over decades. We could also demonstrate, that simple hybridization can be very useful for genotyping or multiplex SNP-detection.
Limitations
Apperently, the PCR has certain limitations compared to other DNA-amplification techniques. A differentiated view reveals that many PCR-associated weak points can be compensated. The choice of appropriate reaction components, the equipment and the overall simplification of the processing allow point-of-care compatible analysis. In addition, the PCR impresses with important characteristics like simple assay design, methodological diversity, multiplexing and excellent specificity. When choosing the right DNA amplification method for the development of an individual rapid test, PCR should not be underestimated.
| Method combined with LFA | Sensitivity | Specificity | Speed | reverse Transcription (one pot) | Robustness | Method. Variety | Simple Assay Design | Multiplexing Ability | Prevention Carryover Cont. |
|---|---|---|---|---|---|---|---|---|---|
| PCR | |||||||||
| LAMP | |||||||||
| RPA | |||||||||
| CRISPR (Label-Sep.) | |||||||||
| CRISPR (Label-Inc.) |