Question: What is the limit of detection of Milenia HybriDetect in pmol?
Over the many years of being in the market with our development tool, we got quite a lot of questions from our customers. Some of these questions are frequently raised. The answers to these questions are given on the HybriDetect Landingpage.
Other questions may be raised individually, but we try to respond to every individual question with the best of our knowledge. If we do not know the answer right now, we run experiments in order to supply our customers with an adequate answer. Just recently a customer wanted to know what the limit of detection of Milenia HybriDetect in pmol is!
Customer reports on the sensitivity of Milenia HybriDetect
To answer this question we did a little literature search and assorted several papers dealing with information about the limit of detection of our HybriDetect.
Piepenburg et al. (2006)
In the very first publication on RPA amplification Piepenburg et al. (2006) used Milenia HybriDetect to detect amplicons. They introduced samples with 10 copies each of MRSAIII, MRSAII and MRSAI and 10.000 copies of MSSA (negative control) as a template in their MRSA assay. The test line was clearly visible on all HybriDetect lateral flow strips with MRSA samples, indicating that the sensitivity of the system must be between 1 and 10 copies of the target gene.
Kiatpathomchai et al.. 2018
In 2008 a paper on “Shrimp Taura syndrome virus detection by reverse transcription loop-mediated isothermal amplification combined with a lateral flow dipstick” was published in the Journal of Virological Methods by Kiatpathomchai et al..
They found out that the combination of their LAMP method and Milenia HybriDetect is overall 100 times more sensitive than an agarose gel and 10 times more sensitive than a nested PCR followed by electrophoresis. Positive side effects of the use of a lateral flow strip were the elimination of a potentially hazardous compound like ethidium bromide, the spimplicity in use and the choice of a true point-of-need application.
Pecchia and Da Lio (2018)
In 2018 Pecchia and Da Lio published a paper with the title “Development of a rapid PCR-Nucleic Acid Lateral Flow Immunoassay (PCR-NALFIA) based on rDNA IGS sequence analysis for the detection of Macrophomina phaseolina in soil”.
The authors found out that the limit of detection of the NALFIA assay was 17.3 fg using 10 μl PCR reaction. In contrast to this the sensitivity of the agarose gel electrophoresis was 17.3 pg using 25 μl of PCR reaction. This means that in the system of Pecchia and Da Lio the sensitivity of Milenia HybriDetect was 1.000 fold higher than the agarose gel.
However, it needs to be stressed that the advantage of Milenia HybriDetect compared to an agarose gel in terms of sensitivity depends on the size of the amplicons to be detected. Pecchia and Da Lio created amplicons of 100 bp length!
The Limit of Detection of HybriDetect can be as low as 0,1 fmol
Beside the literature search, we did experiments to find out the limit of detection of HybriDetect in our lab. Therefore a dual labeled dsDNA (Biotin/FITC), generated by a PCR was transferred to Milenia HybriDetect strips and run. The concentration of the samples ranged from 0 to 250 fmol. The results of the experiment are shown in Figure 1.
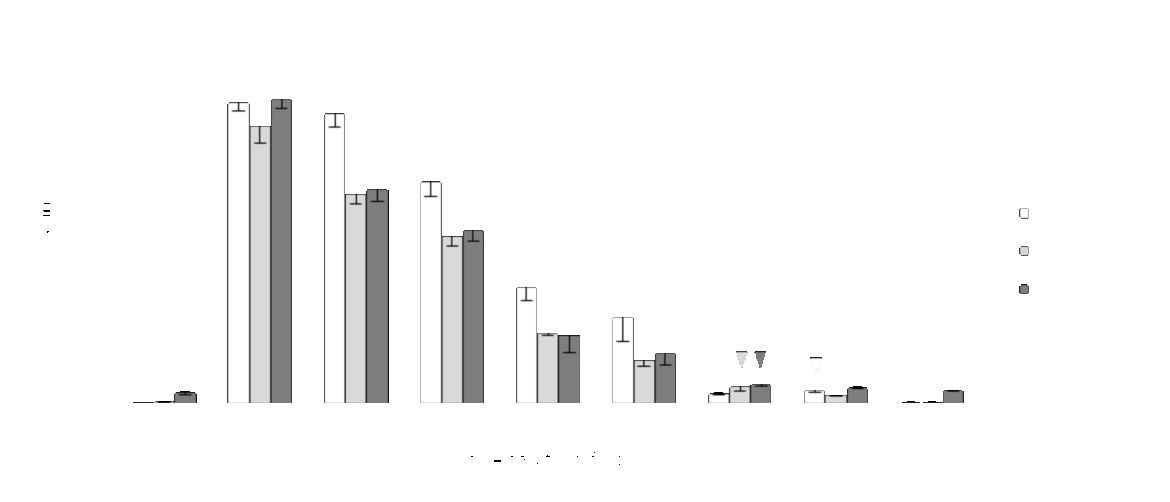
By using dual labeled dsDNA the LOD was calculated below 1 fmol. This approach is limited in terms of precision due to the purification of the PCR fragments, the measurement procedure and the high dilution of the samples.
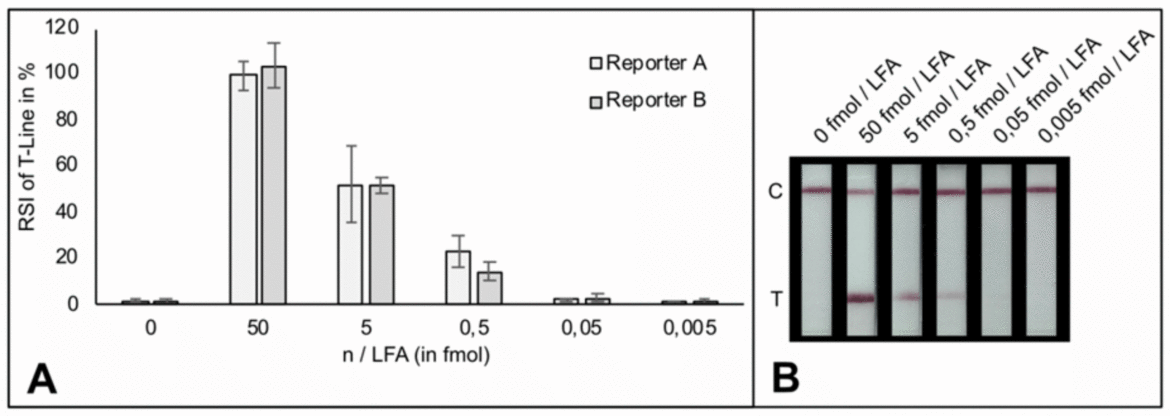
Therefore, a different approach was done and a synthetic reporter gene (dual labeld ssDNA, very short oligos) was tested with HybriDetect. The results indicate that 0,5 fmol of the reporter resulted in clear signals on the T-line of the Milenia HybriDetect test strip. Very slight signals were visible by using the reporter below 0,05 fmol. (Figure 2)
Conclusion
Milenia HybriDetect is a lateral flow test development platform based on a universal test strip. The product is frequently used in molecular biology applications and customers are interested to know how sensitive their assays can be! As a result of a literature search and own experiments it was demonstrated that Milenia HybriDetect could be up to 1.000 times more sensitive than the detection of amplicons via an agarose gel. The sensitivity of HybriDetect could go down as much as 0,05 fmol.
If you would like to know more about our products and news about Milenia Biotec, please follow us on LinkedIn
- Piepenburg O., Williams C.H., Stemple D.L., Armes N.A.: DNA detection using recombination proteins. PLoS Biol 2006; 4(7): e204. DOI:10.1371/journal.pbio.0040204 (2006)
- Kiatpathomchai W., Jaroenram W., Arunrut N., Jitrapakdee S., Flegel TW.: Shrimp Taura syndrome virus detection by reverse transcription loop-mediated isothermal amplification combined with a lateral flow dipstick. Journal of Virological Methods 2008; 153: 214–217 (2008)
- Pecchia S. and Da Lio D.: Development of a rapid PCR-Nucleic Acid Lateral Flow Immunoassay (PCR-NALFIA) based on rDNA IGS sequence analysis for the detection of Macrophomina phaseolina in soil. Journal of Microbiological Mehtods (2018, Vol 151) (2018)