These combined features of sequence specific recognition and cutting have been used for the development of genome editing tools.

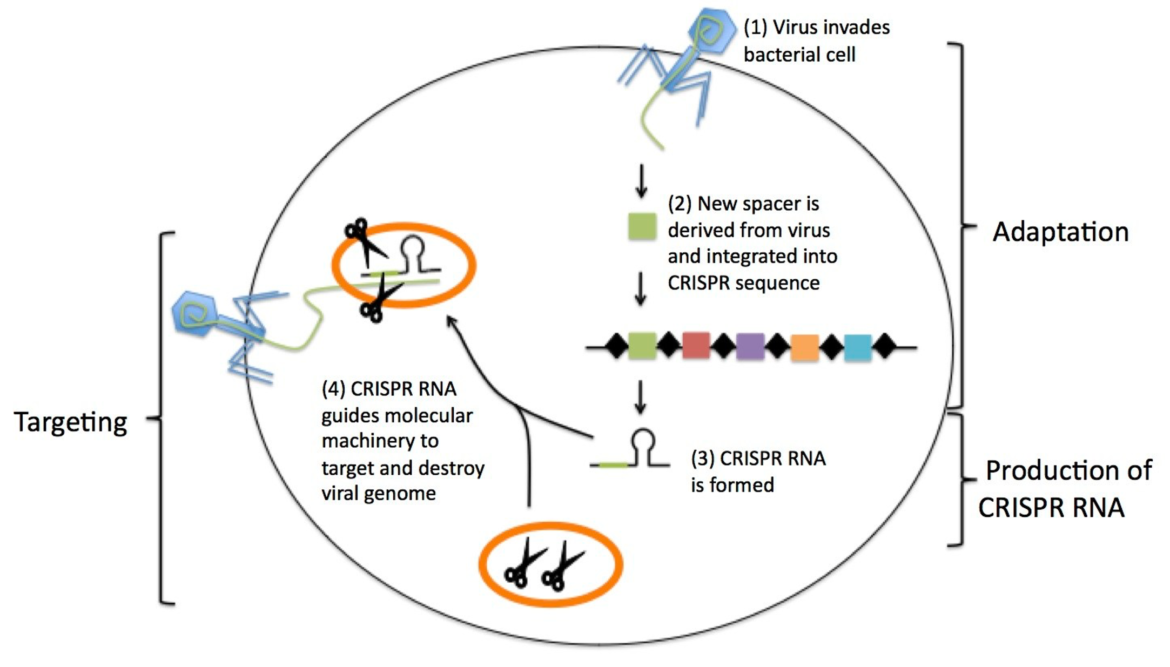
This defense system is based on regions of repeating DNA-sequences, called “Clustered Regularly Interspaced Short Palindromic Repeats, also referred to as CRISPR. In addition, CRISPR-associated proteins (Cas proteins) are required for successful defense. Transcribed CRISPR-RNAs are able to guide Cas protein(s) to the viral genome. A characteristic feature of Cas proteins is the endonuclease activity, which causes the specific degradation of viral nucleic acids. (3, 4)
Importance of Diagnostic Alternatives in the Point-of-Care Field
There is a great need for diagnostic alternatives that are suitable for simple, fast, specific, sensitive, and inexpensive early detection of pathogens. Simple handling and the avoidance of expensive and complex devices are considered to be particularly important, especially for third world countries or regions with limited lab capacities. (6, 7)
This deficiency became very clear in the years 2014 to 2016 during the Ebola outbreak (8). Today we are experiencing an even more extreme situation. The Sars-CoV-2 pandemic affects the whole world and safe, scalable diagnostics is one of the most important issues today. As part of WHO’s response to the outbreak, the R&D Blueprint has been activated to accelerate diagnostics, vaccines and therapeutics for this novel coronavirus (9).
What Makes CRISPR Associated Enzymes Attractive for Diagnostic Purposes?
In recent years, CRISPR/Cas-based detection systems have increasingly come into focus as serious diagnostic alternatives. But why are some Cas proteins particularly suitable for molecular biosensing?
Within the Cas protein classification model, all diagnostically relevant Cas proteins originate from class II. These are multidomain proteins that are guided to the target nucleic acid by associated single stranded RNAs. Another essential feature of diagnostically relevant Cas proteins is a so-called collateral activity, which occurs after a target sequence has been successfully recognized. Single-stranded RNA-/ DNA-fragments are efficiently degraded in a short period of time due to this collateral activity. And this function makes these proteins such interesting tools in diagnostics. By using molecular reporters, the collateral activity can be used for signal generation in a diagnostic approach. Reporters are short DNA- or RNA-fragments that have defined labels at their ends. Reporter cleavage leads to the generation of a detectable signal.
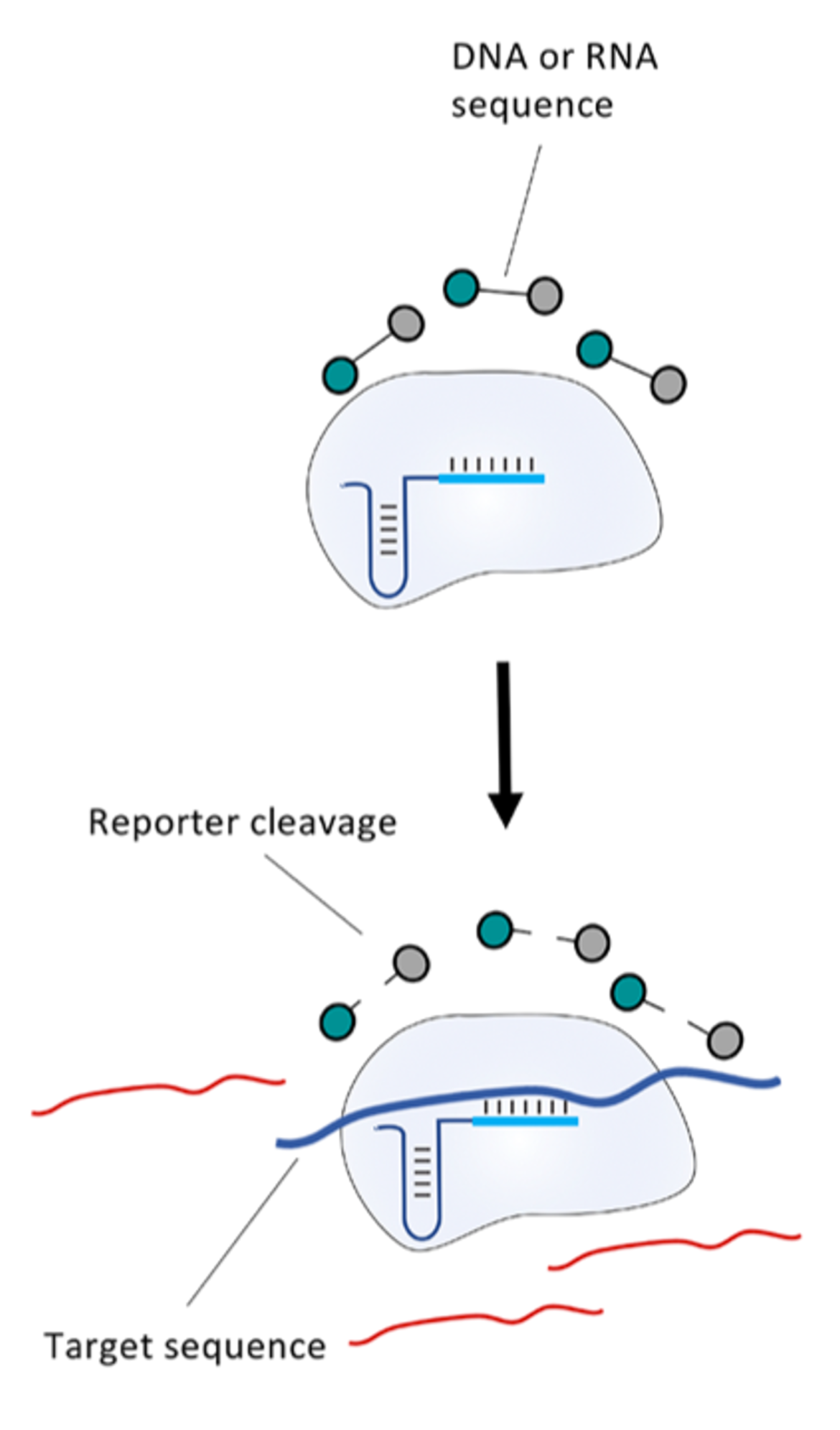
Different Cas proteins have special characteristics and differ in some relevant criteria such as: size, recognition of nucleic acid type, collateral degradation of ssDNA or ssRNA (10). Table 1 gives a brief overview of the three diagnostically relevant Cas proteins and their specific features.
| Characteristics | Cas12a | Cas13 | Cas14 |
|---|---|---|---|
| RNA guided | yes | yes | yes |
| Targeting restrictions | yes (PAM) | weak | yes (PAM) |
| Target type | DNA (ss and ds) | ssRNA | DNA (ss and ds) |
| Classification | Class II, Type V | Class II, Type VI | Class II, Type V |
| Nuclease Acvtivity | ssDNA | ssRNA | ssDNA |
CRISPR/Cas-Systems and Lateral Flow Readout with HybriDetect
CRISPR/Cas-based detection methods can be combined with a simple Lateral Flow Readout. Therefore the universal lateral flow platform HybriDetect is the perfect tool for a sensitive, rapid, equipment-free and simple visualization of test results (11). The general mechanism is explained in the following Figure 3.
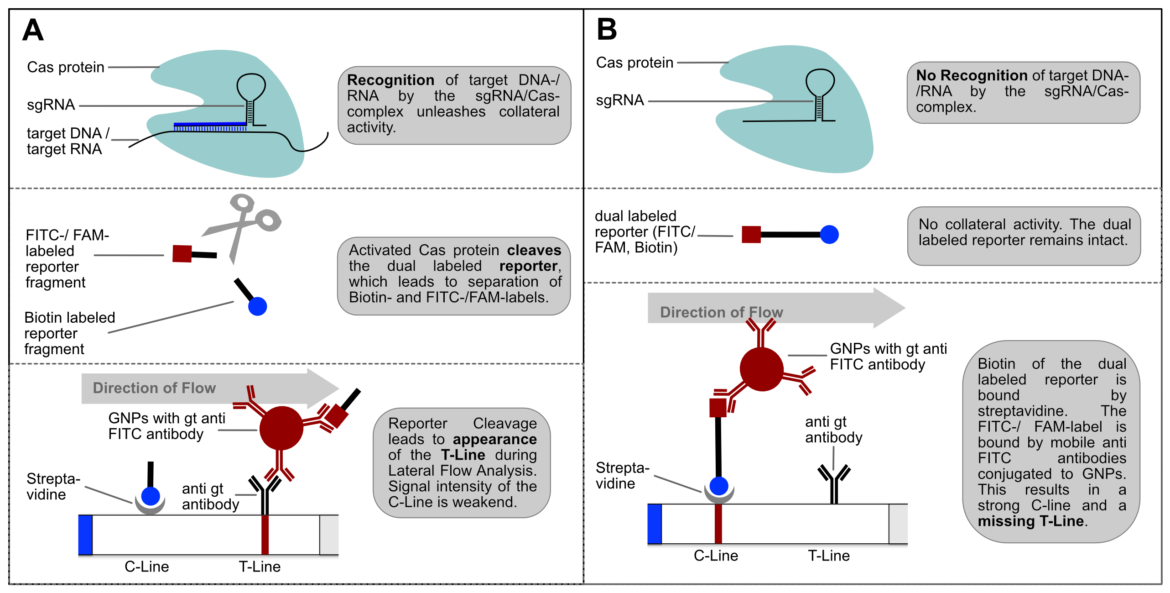
The interpretation of the Milenia HybriDetect dipstick is extremely simple. If the intensity of the T-line exceeds the T-line intensity of the negative control, the test is interpreted as positive. At the same time, the C-line intensity decreases in clear positive results. The simple and intuitive interpretation of the test strips is illustrated in the following figure. About 200 copies of artificial virus RNA can be clearly detected using the CRISPR/Cas-based detection method in combination with HybriDetect lateral flow assay.

CRISPR/Cas-detection methods are mostly combined with pre-amplification steps. Isothermal amplification such as LAMP or RPA are the most frequently used techniques. Combinations of these methods are named SHERLOCK, DETECTR or HOLMES. Recently, it has been proved that these CRISPR/Cas-methods are able to detect pathogenic viral genomes at attomolar levels in a simple, rapid and low-equip Point-of-Care-approach using the Milenia HybriDetect.
- Detection of Zika Virus , SHERLOCK_CRISPR/Cas13-System (12,13)
- Detection of Dengue Virus, SHERLOCK_CRISPR/Cas13-System (12)
- Detection of Human Papillomavirus (HPV) -16 and -18, CRISPR/Cas12a-System (14)
- Detection of SARS-CoV-2, CRISPR/Cas12a-System, CRISPR/Cas13-System (15,16)

- Jinek M, Chylinski K, Fo nfara I, Hauer M, Doudna JA, Charpentier E.: A programmable dual‐RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096): 816‐821. DOI: 10.1126/science.1225829, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6286148/pdf/nihms-995853.pdf ()
- Wright AV, Nuñez JK, Doudna JA: Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell. 2016;164(1‐2):29‐44. DOI: https://doi.org/10.1016/j.cell.2015.12.035 ()
- Singh V, Gohil N, Ramírez García R, Braddick D, Fofié CK: Recent advances in CRISPR‐Cas9 genome editing technology for biological and biomedical investigations. J Cell Biochem. 2018;119(1):81‐94. https://doi.org/10.1002/jcb.26165 ()
- Zetsche B, Gootenberg JS, Abudayyeh OO, et al.: Cpf1 is a single RNA‐guided endonuclease of a class 2 CRISPR‐Cas system. Cell. 2015;163(3):759‐771. Doi: 10.1016/j.cell.2015.09.038.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4638220/pdf/nihms725840.pdf ()
- Barrangou, R. and Marraffini, L.: CRISPR-Cas Systems: Prokaryotes Upgrade to Adaptive Immunity (2014). Molecular Cell 54, 234-244. Original Image: http://sitn.hms.harvard.edu/flash/2014/crispr-a-game-changing-genetic-engineering-technique/ ()
- Zumla A, Al-Tawfiq J, Enne V, Kidd M, Drosten C, Breuer J, Muller M, Hui D, Maeurer M, Bates M, Mwaba P, Al-Hakeem R, Gray G, Gautret P, Al-Rabeeah A, Memish Z, Gant V: Rapid point of care diagnostic tests for viral and bacterial respiratory tract infections-needs, advances, and future prospects. The Lancet Infectious Diseases. 2014 vol: 14 (11) pp: 1123-1135. Doi: 10.1016/S1473-3099(14)70827-8.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7106435/pdf/main.pdf ()
- Chen H, Liu K, Li Z, Wang P.: Point of care testing for infectious diseases.Clinica Chimica Acta. 2019 vol: 493 pp: 138-147. Doi: 10.1016/j.cca.2019.03.008. https://doi.org/10.1016/j.cca.2019.03.008 ()
- Pollock NR, Wonderly B.: Evaluating novel diagnostics in an outbreak setting: lessons learned from Ebola. J Clin Microbiol. 2017;55(5):1255‐1261. https://jcm.asm.org/content/jcm/55/5/1255.full.pdf ()
- WHO: A research and development Blueprint for action to prevent epidemics. Source: https://www.who.int/blueprint/en/ ()
- Aman R, Mahas A, Mahfouz M: Nucleic Acid Detection Using CRISPR/Cas Bio-sensing Technologies. ACS Synthetic Biology. 2020. doi:10.1021/acssynbio.9b00507. https://pubs.acs.org/doi/10.1021/acssynbio.9b00507 ()
- James A, Todd S, Pollak N, Marsh G, Macdonald J: Ebolavirus diagnosis made simple, comparable and faster than molecular detection methods: Preparing for the future.Virology Journal. 2018 vol: 15 (1). https://doi.org/10.1186/s12985-018-0985-8. ()
- Gootenberg J, Abudayyeh O, Kellner M, Joung J, Collins J, Zhang F: Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science vol. 360, pp: 439-444 (2018). doi: 10.1126/science.aaq0179. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5961727/pdf/nihms965860.pdf ()
- Myhrvold C, Freije C, Gootenberg J, Abudayyeh O, Metsky H, Durbin A, Kellner M, Tan A, Paul L, Parham L, Garcia K, Barnes K, Chak B, Mondini A, Nogueira M, Isern S, Michael S, Lorenzana I, Yozwiak N, Macinnis B, Bosch I, Gehrke L, Zhang F, Sabeti P: Field-deployable viral diagnostics using CRISPR-Cas13. Science vol. 360, pp: 444-448 (2018). Doi: 10.1126/science.aas8836. https://science.sciencemag.org/content/360/6387/444/tab-pdf ()
- Tsou J, Leng Q, Jiang F.: A CRISPR Test for Detection of Circulating Nuclei Acids. Translational Oncology. 2019 vol: 12 (12) pp: 1566-1573. doi: 10.1016/j.tranon.2019.08.011 https://www.sciencedirect.com/science/article/pii/S1936523319304140?via%3Dihub ()
- Broughton J, Deng X, Yu G, Fasching C, Streithorst J, Granados A, Sotomayor-Gonzalez A, Gopez A, Hsu E, Gu W, Miller S, Pan C, Wadford D, Chen J, Chiu C, Chiu C: Rapid Detection of 2019 Novel Coronavirus SARS-CoV-2 Using a CRISPR-based DETECTR Lateral Flow Assay. PREPRINT server: bioRxiv. Doi: 10.1101/2020.03.06.20032334. https://www.medrxiv.org/content/10.1101/2020.03.06.20032334v2 ()
- Metsky H, Freije C, Kosoko-Thoroddsen T, Sabeti P, Myhrvold C: CRISPR-based surveillance for COVID-19 using genomically-comprehensive machine learning design. PREPRINT server: bioRxiv. 2020 pp: 2020.02.26.967026. doi: 10.1101/2020.02.26.967026. https://www.biorxiv.org/content/10.1101/2020.02.26.967026v2.full.pdf ()