
Isothermal Amplification and Lateral Flow for the Detection of Pathogenic Nematodes
Root-knot nematodes (Meloidogyne spp.) are able to infect more than 2000 plant species including crops like tomato, potato, carrots, onion and many more. These organisms enter the plant through their root and form abnormal structures called galls, which can lead to a reduction of the overall plant growth (Figure 1). Due to their wide host range and worldwide distribution root-knot nematodes reduce global crop yields by approx. 5%. One of the most important nematode species is Meloidogyne hapla, which is addressed in the following two publications.

Detection of Meloidogyne hapla with LAMP and Lateral Flow
In 2017 Huan Peng and co-workers presented an interesting combination of a FTA-card based DNA extraction method combined with a Loop mediated isothermal amplification (LAMP) and colorimetric and lateral flow readout. LAMP, an isothermal amplification technique, allows sensitive DNA-amplification at 60-65°C. Combination of LAMP with the universal Lateral Flow platform Milenia HybriDetect led to a significant boost in terms of specificity. The authors proved that their diagnostic assay is working in the field to manage one of the destructive nematodes, M. hapla (Figure 2). This paper was published in nature / scienfitic reports (5).

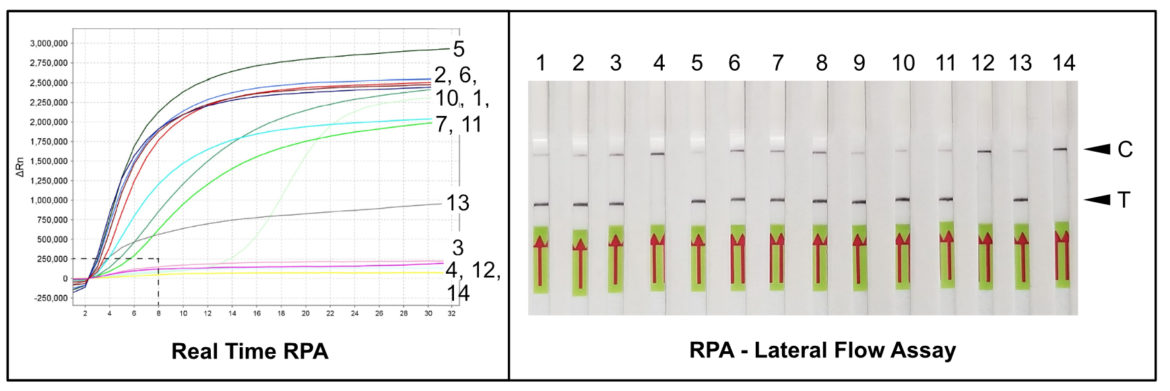
Detection of Meloidogyne hapla with RPA and Lateral Flow
Almost four years later Sergei A. Subbotin and Julie Burbridge from the Plant Pest Diagnostic Center in Sacramento published an alternative field applicable assay for the detection of M. hapla. The authors could benefit from the advantages of another isothermal amplification method, the Recombinase Polymerase Amplification (RPA). The RPA is known as a very robust amplification method and particularly known for its insensitivity to classic inhibitors. In addition, at a constant temperature of 37 – 42 ° C, RPA can efficiently and specifically replicate DNA in few minutes. As a result, the authors were able to show that specific and sensitive detection of M. hapla using crude DNA-extracts is possible in less than 30 minutes (8).
A universal Detection Platform for the Detection of Plant Pathogens
The detection of the root-knot nematode M. hapla is a striking example for the methodological diversity of Nucleic Acid Lateral Flow Immunoassays (NALFIAs). Multiple DNA-amplification methods can be combined with a simple and fast lateral flow readout. Simplification of the sample preparation and amplification reaction in combination with Lateral Flow enables a rapid, robust, field-applicable molecular detection of plant pathogens with a performance comparable to highly sensitive lab-methods, such as RT-qPCR. The use of the universal lateral flow platform Milenia HybriDetect allows a fast, simple and easy-to-adapt NALFIA development.
In recent years there has been an increasing number of publications using the RPA-LFA for point-of-need detection of plant pathogens. In addition to relevant nematodes, bacteria such as Clavibacter michiganensis, viral plant pathogens or oomycetes of the genus Phytophtora were successfully detected with the help of RPA and Lateral Flow (Table 1).
| Plant Pathogen | Description | Host(s) | LFA-compatible amplification method | Reference No. |
|---|---|---|---|---|
| Meloidogyne hapla | Nematode | > 500 plant species (tomato, potato,onions) | LAMP | 5 |
| Meloidogyne hapla | Nematode | > 500 plant species (tomato, potato,onions) | RPA | 8 |
| Phytophtora sojae | Oomycete | Soybean | RPA | 2 |
| Phytophtora hibernalis | Oomycete | Citruses | RPA | 1 |
| Phytophtora infestans | Oomycete | Potato, Tomato | RPA | 4 |
| Clavibater michiganesis subsp. sepedonicus | Bacteria (Actinomycete) | Potato | RPA | 7 |
| Bean Common Mosaic Virus (BCMV) | DNA Virus | Common Bean | RPA | 6 |
| Rice Black-Streakes Dwarf Virus (BPSDV) | RNA Virus | Rice, Maize, Wheat | RT-RPA | 9 |
| Macrophomia phaseolina | Fungus | >500 plant species | PCR | 10 |
Providing sufficient food for the rapidly growing human population is a major challenge for agriculture. High quality detection or monitoring of relevant plant pathogens especially under low resource settings can be a valuable addition in agriculture. To meet this challenge, NALFIAs can make a significant contribution.

- Dai, T., Hu, T., Yang, X., Shen, D., Jiao, B., Tian, W., & Xu, Y.: A recombinase polymerase amplification-lateral flow dipstick assay for rapid detection of the quarantine citrus pathogen in China, Phytophthora hibernalis. PeerJ, 7, e8083. https://doi.org/10.7717/peerj.8083 (2019)
- Dai, T., Yang, X., Hu, T., Jiao, B., Xu, Y., Zheng, X., & Shen, D.: Comparative Evaluation of a Novel Recombinase Polymerase Amplification-Lateral Flow Dipstick (RPA-LFD) Assay, LAMP, Conventional PCR, and Leaf-Disc Baiting Methods for Detection of Phytophthora sojae. Frontiers in microbiology, 10, 1884. https://doi.org/10.3389/fmicb.2019.01884 (2019)
- Jones, J. T., Haegeman, A., Danchin, E. G., Gaur, H. S., Helder, J., Jones, M. G., Kikuchi, T., Manzanilla-López, R., Palomares-Rius, J. E., Wesemael, W. M., & Perry, R. N.: Top 10 plant-parasitic nematodes in molecular plant pathology. Molecular plant pathology, 14(9), 946–961. https://doi.org/10.1111/mpp.12057 (2013)
- Lu, X., Zheng, Y., Zhang, F., Yu, J., Dai, T., Wang, R., Tian, Y., Xu, H., Shen, D., & Dou, D.: A Rapid, Equipment-Free Method for Detecting Phytophthora infestans in the Field Using a Lateral Flow Strip-Based Recombinase Polymerase Amplification Assay. Plant disease, 104(11), 2774–2778. https://doi.org/10.1094/PDIS-01-20-0203-SC (2020)
- Peng, H., Long, H., Huang, W., Liu, J., Cui, J., Kong, L., Hu, X., Gu, J., & Peng, D.: Rapid, simple and direct detection of Meloidogyne hapla from infected root galls using loop-mediated isothermal amplification combined with FTA technology. Scientific reports, 7, 44853. https://doi.org/10.1038/srep44853 (2017)
- Qin, J., Yin, Z., Shen, D. et al.: Development of a recombinase polymerase amplification combined with lateral flow dipstick assay for rapid and sensitive detection of bean common mosaic virus. Phytopathol Res 3, 3 (2021). https://doi.org/10.1186/s42483-021-00080-3 (2021)
- Sagcan, H., & Turgut Kara, N.: Detection of Potato ring rot Pathogen Clavibacter michiganensis subsp. sepedonicus by Loop-mediated isothermal amplification (LAMP) assay. Scientific reports, 9(1), 20393. https://doi.org/10.1038/s41598-019-56680-9 (2019)
- Subbotin, S. A., & Burbridge, J.: Sensitive, Accurate and Rapid Detection of the Northern Root-Knot Nematode, Meloidogyne hapla, Using Recombinase Polymerase Amplification Assays. Plants (Basel, Switzerland), 10(2), 336. https://doi.org/10.3390/plants10020336 (2021)
- Zhao, C., Sun, F., Li, X., Lan, Y., Du, L., Zhou, T., & Zhou, Y.: Reverse transcription-recombinase polymerase amplification combined with lateral flow strip for detection of rice black-streaked dwarf virus in plants. Journal of virological methods, 263, 96–100. https://doi.org/10.1016/j.jviromet.2018.11.001 (2018)
- Pecchia, S., & Da Lio, D.: Development of a rapid PCR-Nucleic Acid Lateral Flow Immunoassay (PCR-NALFIA) based on rDNA IGS sequence analysis for the detection of Macrophomina phaseolina in soil. Journal of microbiological methods, 151, 118–128. https://doi.org/10.1016/j.mimet.2018.06.010 (2018)

